C++ Example Structures.C
ABOUT THIS EXAMPLE:
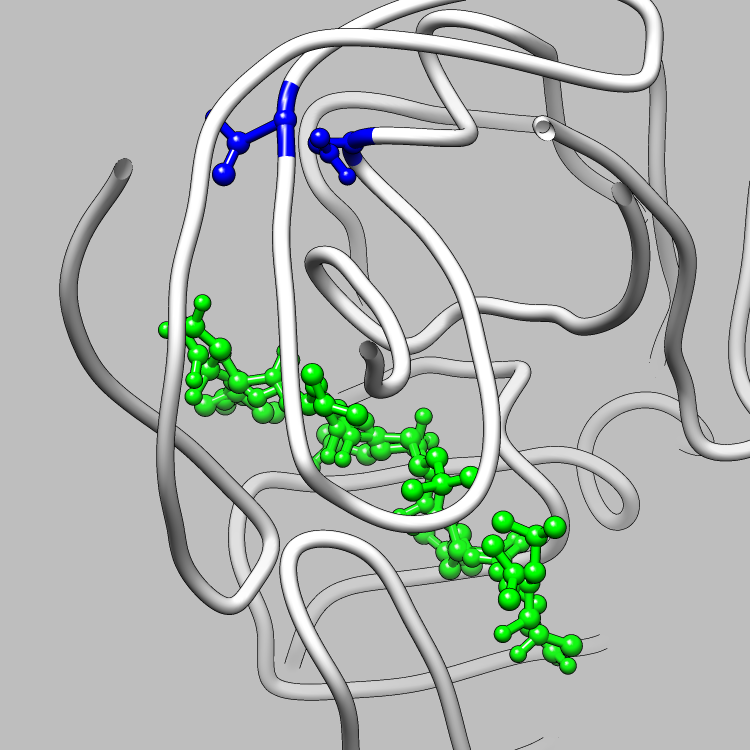
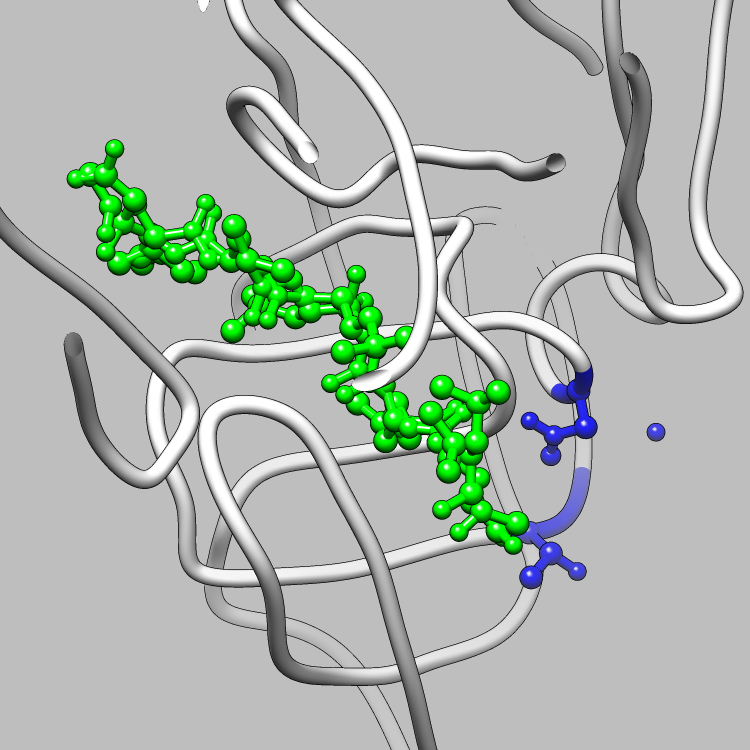
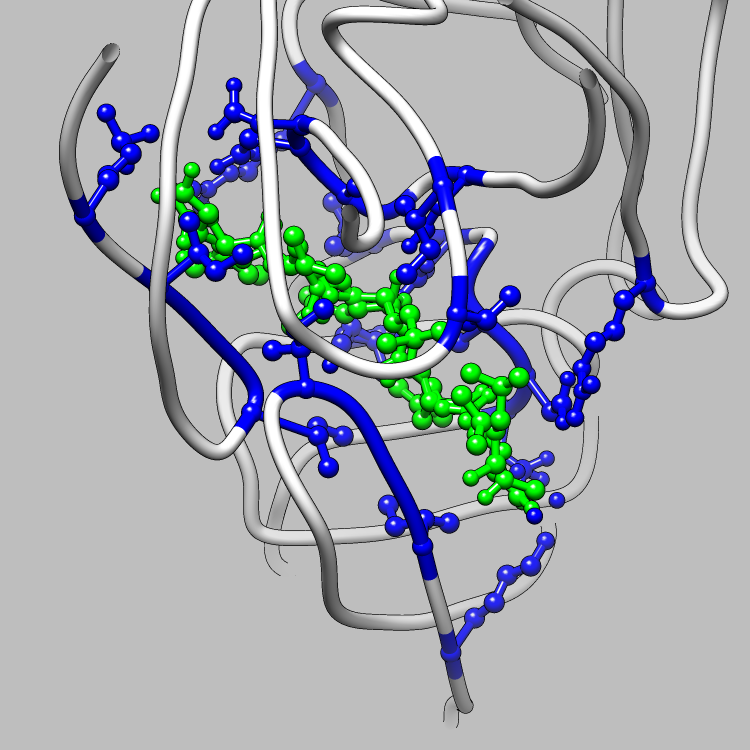
This example is one of a few that shows how the CIFPARSE-OBJ library can be used to interface with Chimera, such that useful and interesting aspects of a molecule, obtainable via parsing CIF files, can be located and used as the subject of a Chimera render or animation. This particular example shows how to retrieve and iterate over the struct_site_gen category, which delineates members of structurally relevant sites in a molecule, and locate all structurally relevant sites for Chimera to emphasize and animate.
Build Instructions:
Files: Structures.C, 5HVP.cif, Structures.sh
Save Structures.C to /path/to/cifparse-obj-vX.X-prod-src/parser-test-app-vX.X/src/ Save the CIF file anywhere, e.g., /path/to/cifparse-obj-vX.X-prod-src/parser-test-app-vX.X/bin/ Add Structures.ext to the BASE_MAIN_FILES list in the Makefile in /path/to/cifparse-obj.vX.X-prod-src/parser-test-app-vX.X Execute make in the same directory as the Makefile cd to bin, where the executable has been made, and run ./Structures /path/to/5HVP.cif, which generates a /path/to/5HVP.com file which you can open with chimera /path/to/5HVP.com Alternatively, you can save the script to /path/to/cifparse-obj-vX.X-prod-src/parser-test-app-vX.X/bin/, set the Chimera path, and run ./Structures.sh /path/to/5HVP.cif, which will automate the process
Functions to Note
#include "CifFile.h"
- string CifFile::GetFirstBlockName() ►
- Returns the first data block name. CifFile inherits this method from TableView. Related: CifFile::GetBlockNames(vector<string>& blockNames).
- Block& CifFile::GetBlock(const string& blockName) ►
- Retrieves a data block specified by some blockName. CifFile inherits this method from TableView.
- ISTable& Block::GetTable(const string& name) ►
- Retrieves a table (i.e., category) within the block, specified by some name.
#include "ISTable.h"
- unsigned int ISTable::GetNumRows() ►
- Returns the numbers of rows in the table (i.e., category).
- const string& operator()(const unsigned int rowIndex, const string colName) ►
- Returns the value of the attribute colName at row index rowIndex
Molecular Graphics of the Structurally Relevant Sites in 5HVP
(LIGAND IN GREEN)
/*************************
* Structures.C
*
* For some CIF file, generate a Chimera command (COM) file
* to iterate through and emphasize each structurally relevant site.
*
* Lines with superscriptions contain footnoted references or explanations.
*************************/
#include <cstring>
#include <fstream>
#include <iostream>
#include <map>
#include <string>
#include <vector>
#include "CifFile.h"
#include "CifParserBase.h"
#include "ISTable.h"
void prepareOutFile(std::ofstream& outFile, const string& cifFileName);
void showUsage();
void writeSite(std::ofstream& outFile, const string& select);
int main(int argc, char **argv)
{
if (argc != 2)
{
showUsage();
}
// The name of the CIF file
string cifFileName = argv[1];
// A string to hold any parsing diagnostics
string diagnostics;
// Create CIF file and parser objects
CifFile *cifFileP = new CifFile;
CifParser *cifParserP = new CifParser(cifFileP);
// Parse the CIF file
cifParserP->Parse(cifFileName, diagnostics);
// Delete the CIF parser, as it is no longer needed
delete cifParserP;
// Display any diagnostics
if (!diagnostics.empty())
{
std::cout << "Diagnostics: " << std::endl << diagnostics << std::endl;
}
// Get the first data block name in the CIF file
string firstBlockName = cifFileP->GetFirstBlockName();
// Retrieve the first data block
Block &block = cifFileP->GetBlock(firstBlockName);
// Retrieve the table corresponding to the struct_site_gen category, which delineates structurally relevant sites1
ISTable& struct_site_gen = block.GetTable("struct_site_gen");
// Use the CIF file name to generate the Chimera command file (.COM) name
size_t fileExtPos = cifFileName.find(".cif");
string outFileName = cifFileName.substr(0, fileExtPos) + ".com";
// Create the command file
std::ofstream outFile;
outFile.open(outFileName.c_str());
// Write out some basic Chimera initialization commands
prepareOutFile(outFile, cifFileName);
// A string to remember the ID of the current site being read
string currentSite;
// A Chimera selection string for the site members
string select;
// Iterate through every row in the struct_site_gen category table
for (unsigned int i = 0; i < struct_site_gen.GetNumRows(); ++i)
{
// Get the site identifier for the current row
string site_id = struct_site_gen(i, "site_id");
// Check for the first site
if (currentSite.empty())
{
currentSite = site_id;
}
// Check for a new site
else if (currentSite != site_id)
{
// Write out commands for Chimera to customize the display of the current site
writeSite(outFile, select);
// Clear the Chimera selection string
select.clear();
// Make this site our new current site
currentSite = site_id;
}
// Otherwise, we are adding another site member
else
{
select += " | ";
}
// Retrieve all information necessary to uniquely identify this site member2
string asym_id = struct_site_gen(i, "auth_asym_id");
string comp_id = struct_site_gen(i, "auth_comp_id");
string seq_id = struct_site_gen(i, "auth_seq_id");
// Add the member to the Chimera selection string for this site3
select += ":" + seq_id + "," + comp_id + "." + asym_id;
}
// Write out the last site
writeSite(outFile, select);
// Write out the Chimera close command
outFile << "stop\n";
// Close the COM file as all sites have been processed
outFile.close();
return 0;
}
void prepareOutFile(std::ofstream& outFile, const string& cifFileName)
{
outFile << "windowsize 500 500\n"; // Set the window size to 500 x 500 px
outFile << "open " + cifFileName << std::endl; // Open the CIF file
outFile << "preset apply pub 4\n"; // Apply publication preset #4
outFile << "color white\n"; // Color the entire molecule white
outFile << "set bg_color gray\n"; // Color the background gray
outFile << "disp; repr bs; set silhouette\n"; // Represent the atoms in ball-and-stick format
outFile << "savepos fullview\n"; // Remember this position (i.e., the full view of molecule)
}
void showUsage()
{
std::cout << "Usage: ./Structures /path/to/file.cif" << std::endl;
exit(1);
}
void writeSite(std::ofstream& outFile, const string& select)
{
outFile << "color green ligand\n"; // Color the ligand green
outFile << "sel " + select << std::endl; // Select the site members
outFile << "color blue sel\n"; // Color them blue
outFile << "sel sel | ligand; wait 20\n"; // Further select the ligand
outFile << "focus sel; wait 25; ~disp ~sel; wait 100\n"; // Focus in on the selection and hide non-selected atoms
outFile << "disp ~sel; reset fullview 20\n"; // Return to the full view of the molecule
outFile << "color white sel; ~sel; wait 20\n"; // Uncolor and drop the selection
}